Chapter 7 - Spatial Summary Tools
2025-03-27
Source:vignettes/articles/2.7-SummaryTools.Rmd
2.7-SummaryTools.RmdChapter 7: Spatial Summary Tools
Authors: Dimitrios Markou, Danielle Ethier
In Chapter 6, you downloaded satellite imagery from the Copernicus SENTINEL-2 mission and calculated spectral indices (NDWI, NDVI) over La Mauricie National Park to combine with NatureCounts data from the Quebec Breeding Bird Atlas (2010 - 2014). In this chapter, you will create data summaries and visualizations using NatureCounts data and environmental covariates.
This chapter uses the data products prepared in Chapters 4-6, and the National Park boundary and NatureCounts data downloaded in section 4.1 Data Setup from Chapter 4: Elevation Data. For quick access, all data are available for download via the Google Drive data folder. If you wish to gain experience in how to download, process, and save the environmental layers yourself, return to the earlier chapters of this tutorial series and explore the Additional Resources articles.
7.0 Learning Objectives
By the end of Chapter 7 - Spatial Summary Tools, users will know how to:
- Explore variable relationships and data distribution: Diagnostics Plots
- Show the relative abundance (number of individuals) of a species in a community: Species Rank Plots
- Explore the relationship of species richness with environmental variables: Regression Plots
- Visualize species-specific distributions: Presence/Absence Plots
Load the required packages:
library(tidyverse)
library(sf)
library(MASS)
library(corrr)
library(ggcorrplot)
library(FactoMineR)
library(factoextra)
library(ggplot2)
library(naturecounts)
library(tidyr)This tutorial uses environmental data extracted or calculated from
previous chapters of the Spatial Data Tutorial. In Chapter 5: Land Cover Data, the
landscapemetrics package was used to calculate some common
metrics that help quantify habitat composition and configuration:
- Percent landcover (PLAND) - percent of landscape of a given class
- Edge density (ED) - total boundary length of all patches of a given class per unit area
- Number of patches (NP) - number of unique contiguous units
In Chapter 6: Satellite Imagery, the Normalized Difference Vegetation Index (NDVI) was calculated using Sentinel-2 imagery and custom functions .
7.1 Data Setup
In Chapter 4, Chapter 5, and Chapter 6 you extracted elevation,
land cover, and NDVI values, respectively, at surveys sites across La
Mauricie National Park. These data were uploaded to the Google
Drive for your convenience. These data should be neatly stored in
your env_covariates subdirectory.
Run the code chunk below to create your subdirectory, if necessary.
if (!dir.exists("data/env_covariates")) { # checks if "env_covariates" subdirectory exists
print("Subdirectory does not exist. Creating now...")
dir.create("data/env_covariates", recursive = TRUE) # if not, creates subdirectory
} else {
print("Subdirectory already exists.")
}
#> [1] "Subdirectory already exists."Let’s download all the environmental covariates and join them to a common dataframe.
# List the dataframes
env_covariates <- list.files(path = here::here("misc/data/env_covariates"), # drop `here::here()` and paste appropriate path to your data
pattern = "\\.csv$",
full.names = TRUE)
# Read each CSV into a list of dataframes
env_covariates_list <- lapply(env_covariates, read_csv)
# Combine NatureCounts and environmental covariates
env_covariates_df <- Reduce(function(x, y) left_join(x, y, by = "point_id"), env_covariates_list)Read in the NatureCounts data you downloaded from Chapter 4: Elevation Data or the Google
Drive. This file should be in your data folder.
Create an sf object from the NatureCounts data that
represents the unique point count locations.
mauricie_birds_sf <- mauricie_birds_df %>%
st_as_sf(coords = c("longitude", "latitude"), crs = 4326) Select attribute columns for better readability. Explicitly call the
dplyr function to avoid namespace clashes.
mauricie_birds_sf <- mauricie_birds_sf %>%
dplyr::select(SiteCode, survey_year, survey_month, survey_day, english_name, ObservationCount, geometry)Assign a point identifier to each location based on its unique geometry.
mauricie_birds_summary <- mauricie_birds_sf %>%
group_by(SiteCode, geometry) %>%
mutate(point_id = cur_group_id()) %>%
ungroup() %>%
distinct() %>%
st_drop_geometry(mauricie_birds_summary) # drops geometry and converts to regular dataframeCalculate the species diversity and abundance at each point.
biodiversity_count <- mauricie_birds_summary %>%
group_by(point_id) %>%
summarise(n_species = n_distinct(english_name),
n_individuals = sum(ObservationCount, na.rm = TRUE), .groups = "drop") # Count unique speciesJoin the env_covariates_df and
biodiversity_count dataframes. Cleanup variable names.
7.2 Diagnostic Plots
Diagnostic plots like summaries, scatterplot matrices, and histograms are essential for evaluating variable relationships, detecting collinearity, and understanding data distributions. They help you understand your data by identifying patterns, outliers, and multicollinearity, ensure accurate model assumptions, and improve data-driven decision-making in future statistical analyses.
Retrieve basic summary info for the environmental data.
summary(enviro_data_df)
#> point_id elevation ed np pland_needleleaf pland_broadleaf pland_mixed_forest pland_shrubland
#> Min. : 1.00 Min. :110.8 Min. :104.8 Min. :158.0 Min. : 3.786 Min. : 0.2404 Min. :33.86 Min. :0.02405
#> 1st Qu.: 60.75 1st Qu.:209.2 1st Qu.:123.6 1st Qu.:202.0 1st Qu.: 9.579 1st Qu.: 4.1029 1st Qu.:49.23 1st Qu.:0.22852
#> Median :120.50 Median :266.6 Median :134.5 Median :236.5 Median :18.256 Median : 9.1959 Median :56.54 Median :0.52980
#> Mean :120.50 Mean :271.6 Mean :134.6 Mean :245.0 Mean :20.410 Mean :12.1587 Mean :54.79 Mean :1.31667
#> 3rd Qu.:180.25 3rd Qu.:343.6 3rd Qu.:143.9 3rd Qu.:287.0 3rd Qu.:28.252 3rd Qu.:20.4434 3rd Qu.:61.10 3rd Qu.:2.05114
#> Max. :240.00 Max. :426.9 Max. :168.7 Max. :369.0 Max. :55.681 Max. :45.2773 Max. :76.11 Max. :5.93750
#>
#> pland_grassland pland_wetland pland_barren pland_urban pland_water ndvi n_species n_individuals
#> Min. :0.01202 Min. :0.01201 Min. :0.01202 Min. :0.01203 Min. : 0.6377 Min. :0.03471 Min. : 1.000 Min. : 1.00
#> 1st Qu.:0.09623 1st Qu.:0.01204 1st Qu.:0.01203 1st Qu.:0.11729 1st Qu.: 4.8373 1st Qu.:0.44175 1st Qu.: 5.000 1st Qu.: 6.00
#> Median :0.22842 Median :0.03602 Median :0.01203 Median :0.42151 Median : 9.4500 Median :0.54457 Median : 8.000 Median :11.00
#> Mean :0.24828 Mean :0.03624 Mean :0.01303 Mean :0.56084 Mean :10.5240 Mean :0.50169 Mean : 9.108 Mean :13.93
#> 3rd Qu.:0.34302 3rd Qu.:0.05112 3rd Qu.:0.01204 3rd Qu.:0.61389 3rd Qu.:13.5206 3rd Qu.:0.60198 3rd Qu.:11.250 3rd Qu.:19.00
#> Max. :0.67356 Max. :0.09627 Max. :0.02404 Max. :6.62260 Max. :32.1897 Max. :0.66887 Max. :24.000 Max. :47.00
#> NA's :17 NA's :164 NA's :228Create a scatterplot matrix to assess the collinearity of the variables.
pairs(~ n_species + n_individuals + elevation + np + ed + pland_mixed_forest + ndvi, data=enviro_data_df,
main="Scatterplot Matrix of Environmental Variables")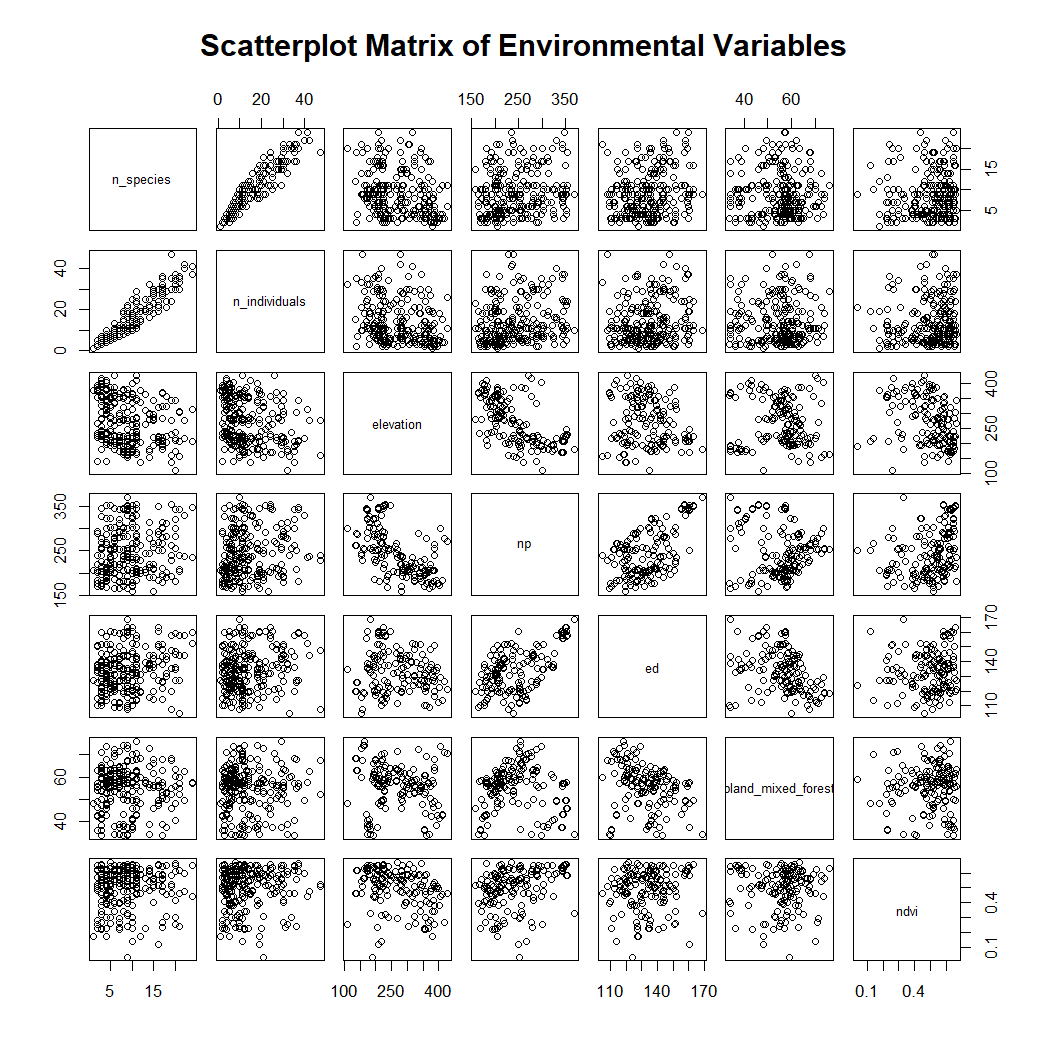
Inspect the distribution and ‘skewness’ of the environmental data using a histogram plot.
# Select variables and pivot longer for faceting
hist_data <- enviro_data_df %>%
dplyr::select(elevation, ed, pland_mixed_forest, ndvi) %>%
pivot_longer(cols = everything(), names_to = "variable", values_to = "value")
# Create faceted histogram plot
ggplot(hist_data, aes(x = value)) +
geom_histogram(bins = 10, fill = "skyblue", color = "black") +
facet_wrap(~ variable, scales = "free") + # Separate histograms for each variable
theme_minimal() +
labs(title = "Multipanel Histogram of Environmental Variables",
x = "Value",
y = "Frequency")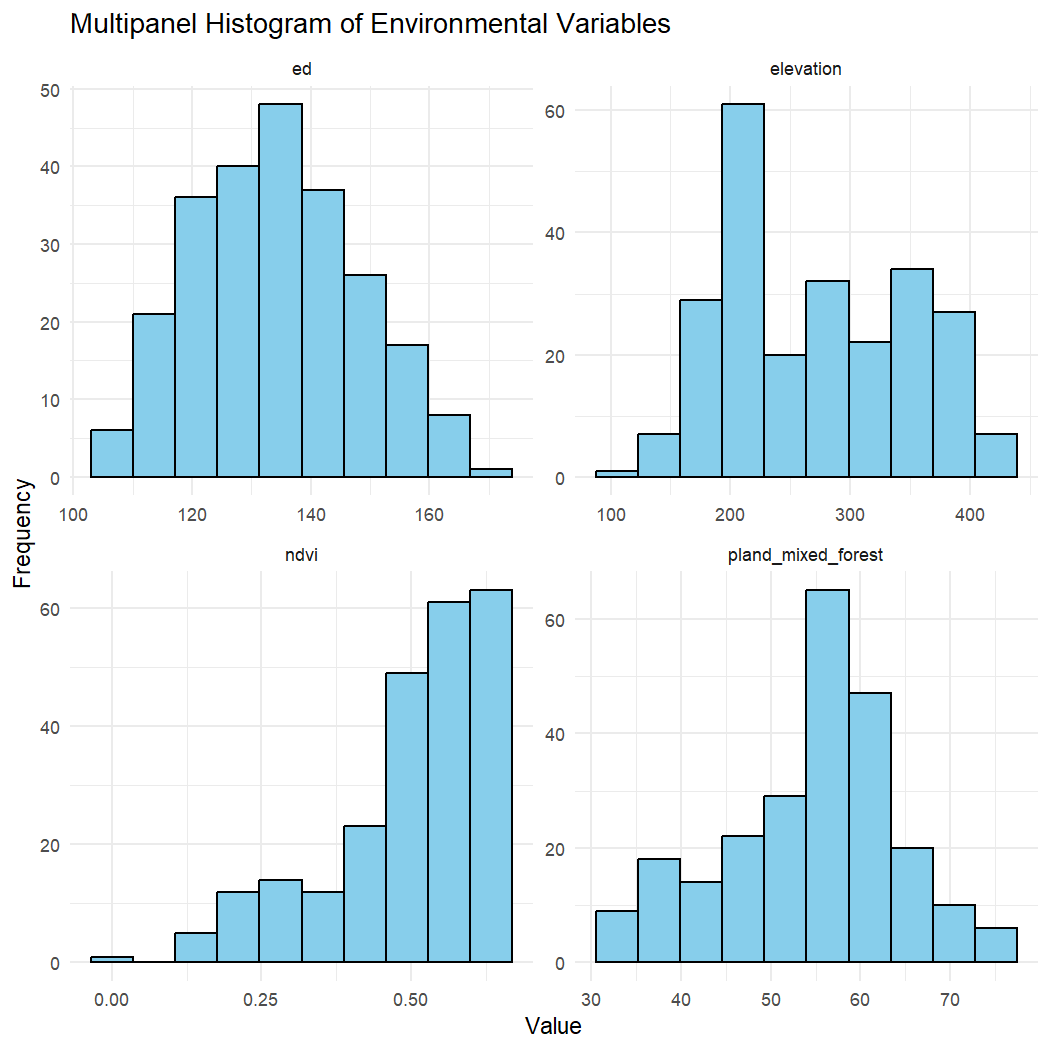
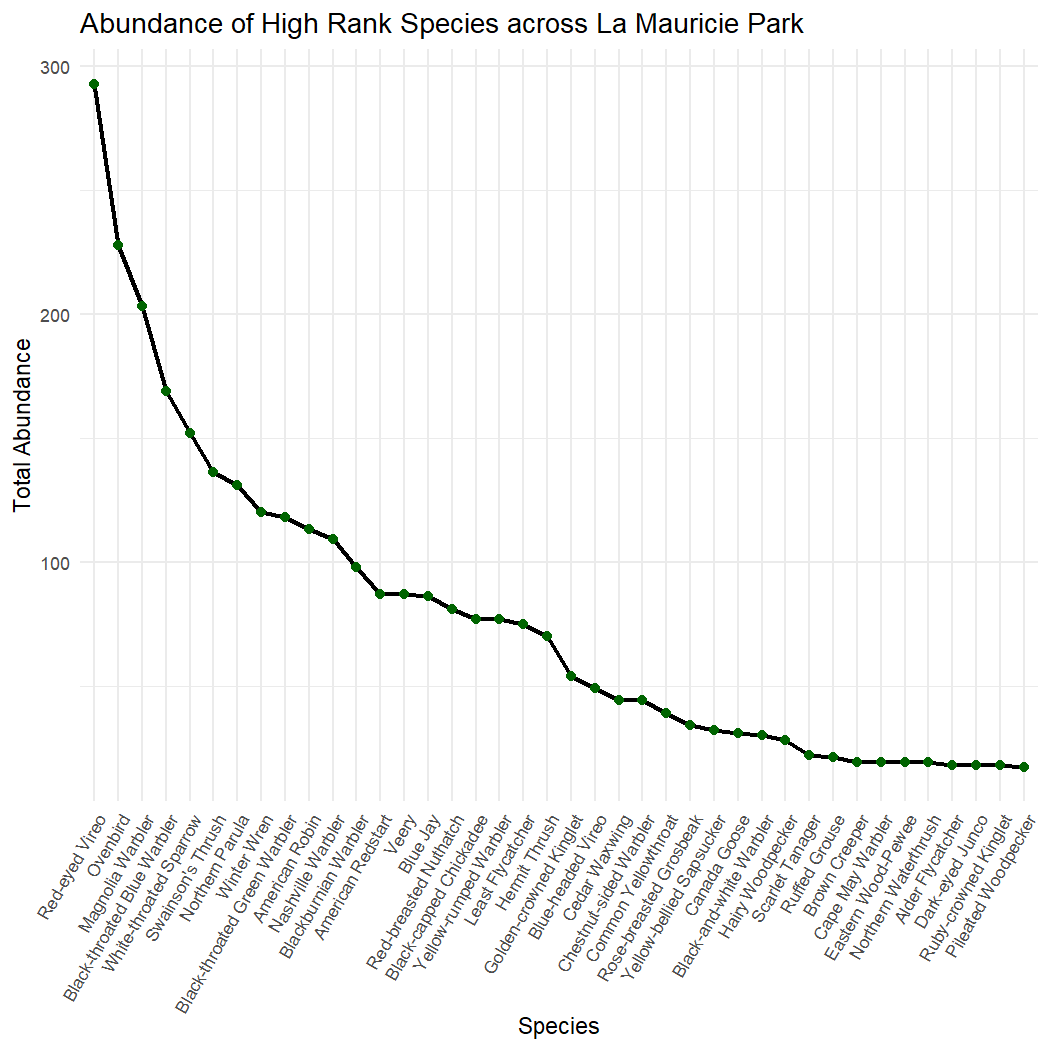
7.3 Species Rank Plots
Species rank plots show the relative abundance (number of individuals) of a species in a community. The number of individuals of each species are sorted in ascending or descending order.
Group the NatureCounts data by species and rank them in order of abundance.
species_rank <- mauricie_birds_summary %>%
filter(!is.na(english_name)) %>% # Remove rows with NA in english_name
group_by(english_name) %>% # Group by species only
summarize(total_abundance = sum(ObservationCount, na.rm = TRUE), .groups = "drop") %>%
arrange(desc(total_abundance)) %>% # Sort in descending order of abundance
slice_max(total_abundance, n = 40) %>% # Keep only the top 40 species
mutate(rank = row_number()) # Assign rank to each speciesPlot the abundance of each species and its rank across the entire park.
ggplot(species_rank, aes(x = reorder(english_name, rank), y = total_abundance)) +
geom_line(group = 1, size = 1, color = "black") +
geom_point(size = 2, color = "darkgreen") +
theme_minimal() +
labs(
title = "Abundance of High Rank Species across La Mauricie Park",
x = "Species",
y = "Total Abundance"
) +
theme(
axis.text.x = element_text(size = 8, angle = 60, hjust = 1)
)
7.4 Regression Plots
Create scatter plots with regression lines to explore the relationship of species richness with each environmental variable.
# Elevation vs. n_species
ggplot(enviro_data_df, aes(x = elevation, y = n_species)) +
geom_point() +
geom_smooth(method = "lm", col = "blue") +
labs(title = "Relationship between Elevation and Species Richness")
#> `geom_smooth()` using formula = 'y ~ x'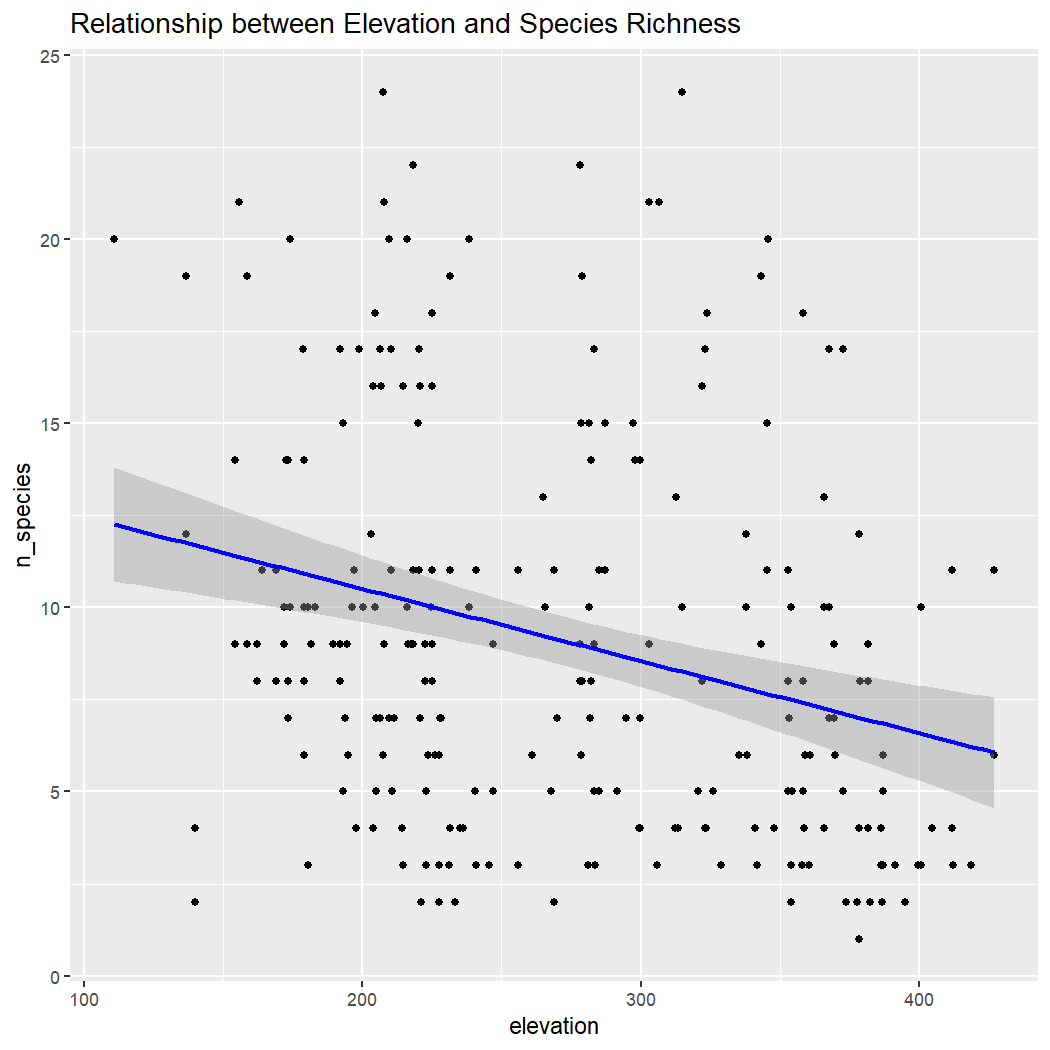
# PLAND vs. n_species
ggplot(enviro_data_df, aes(x = pland_mixed_forest, y = n_species)) +
geom_point() +
geom_smooth(method = "lm", col = "blue") +
labs(title = "Relationship between PLAND and Species Richness")
#> `geom_smooth()` using formula = 'y ~ x'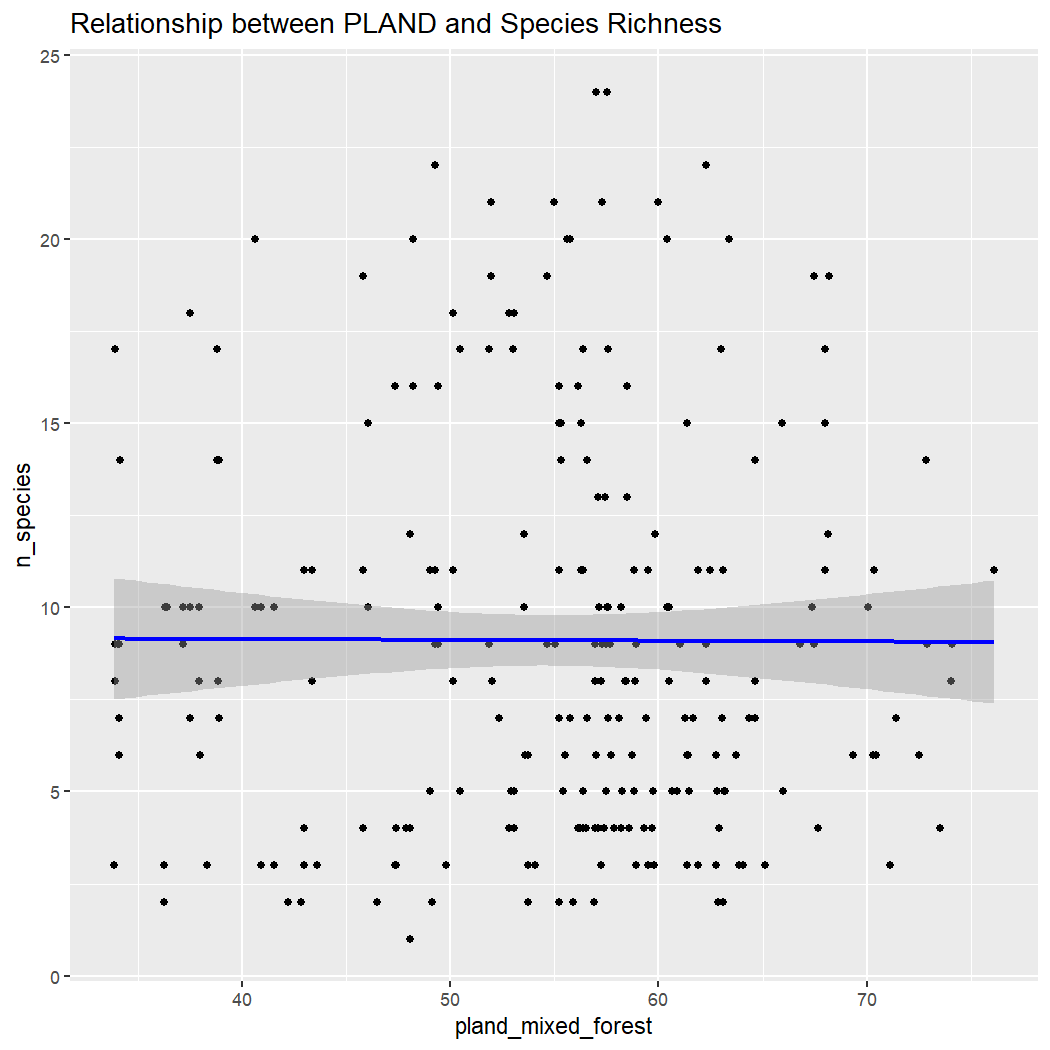
# Edge Density vs. n_species
ggplot(enviro_data_df, aes(x = ed, y = n_species)) +
geom_point() +
geom_smooth(method = "lm", col = "blue") +
labs(title = "Relationship between Edge Density and Species Richness")
#> `geom_smooth()` using formula = 'y ~ x'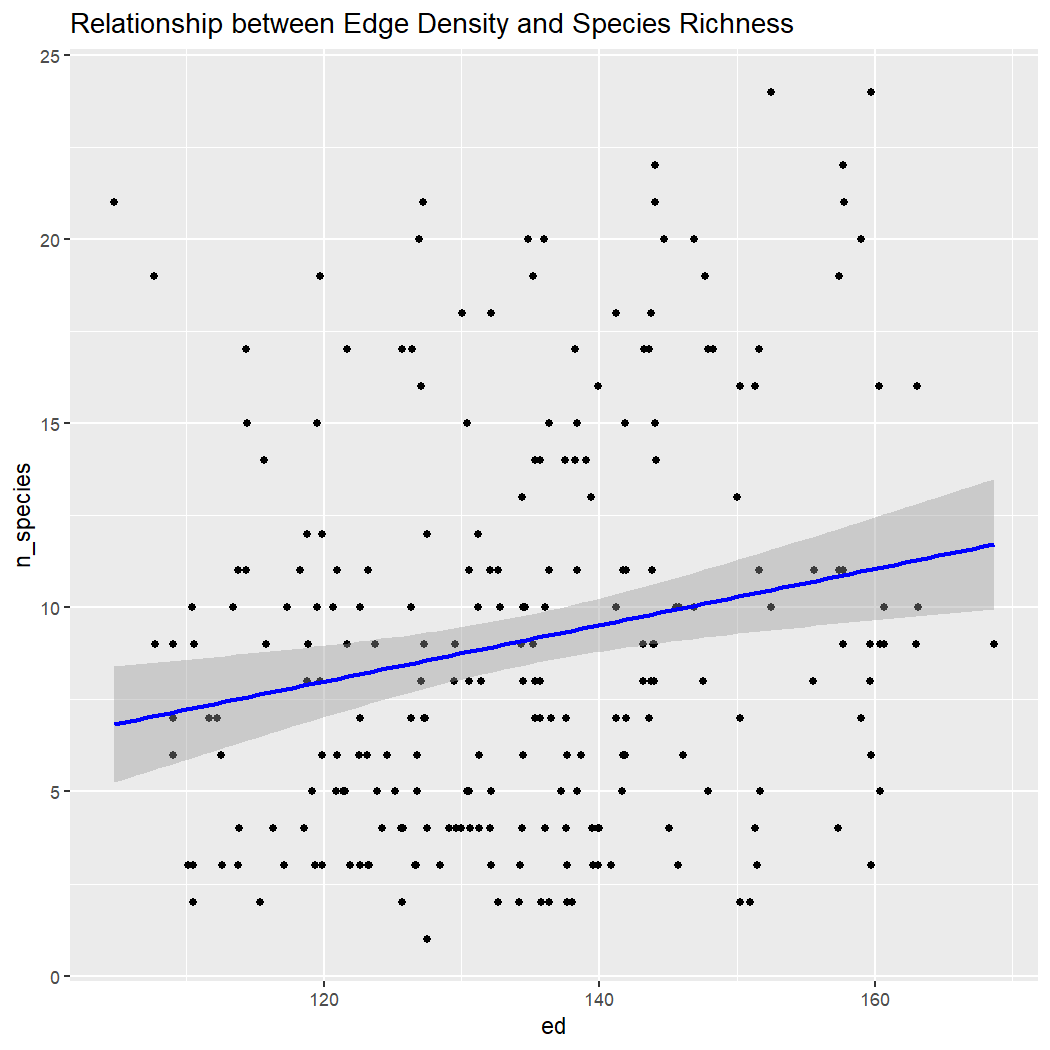
# NDVI vs. n_species
ggplot(enviro_data_df, aes(x = ndvi, y = n_species)) +
geom_point() +
geom_smooth(method = "lm", col = "blue") +
labs(title = "Relationship between NDVI and Species Richness")
#> `geom_smooth()` using formula = 'y ~ x'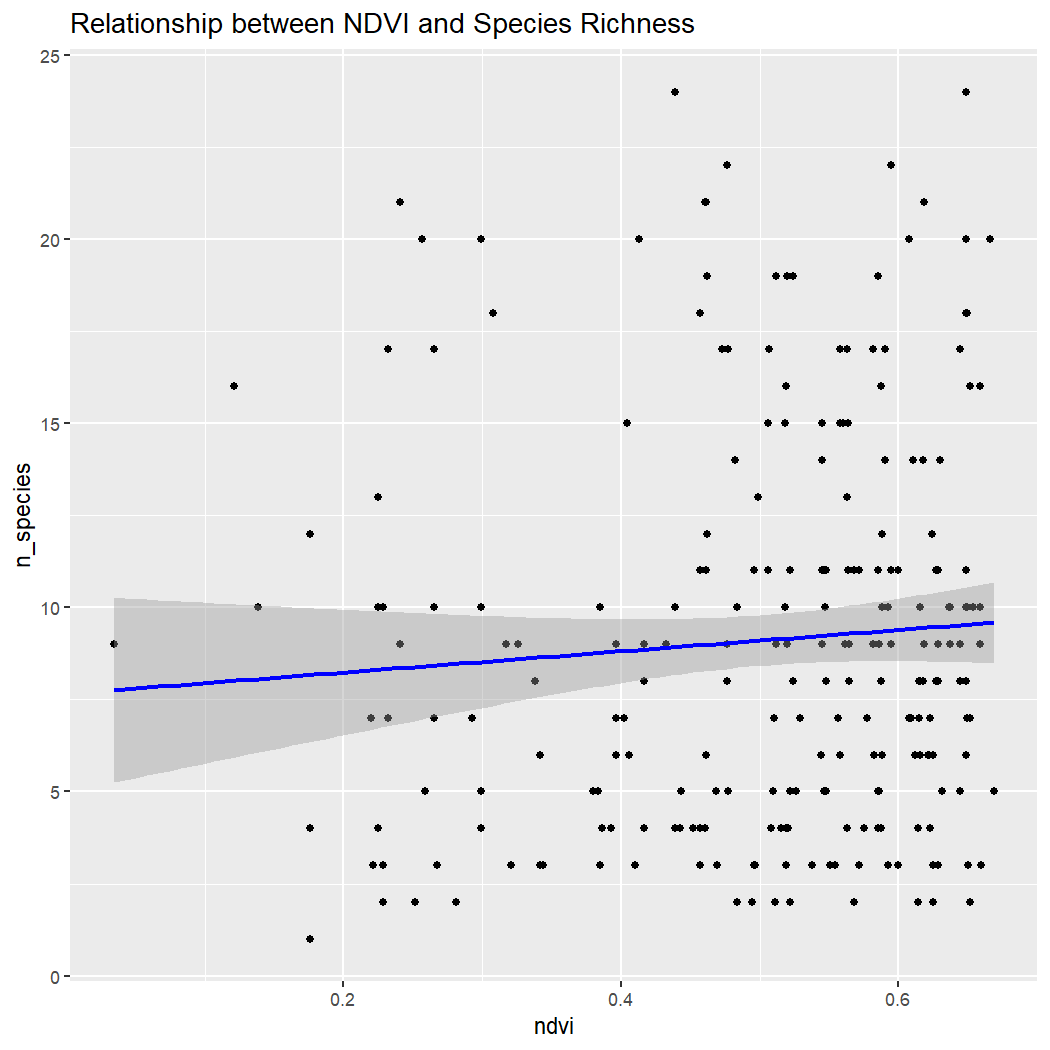
Conclusion: There is no clear relationship between species richness and elevation, ED, or NDVI within La Mauricie National Park. Note: These results assume that sampling is random with regards to elevation, which is unlikely because sampling is done along roads which tend to be at lower elevations.
7.5 Presence/Absence
Create an events matrix containing the geometry for each unique survey event location.
Recall that most NatureCounts datasets will have a unique SiteCode for each survey point. However, this is not the case the the Quebec Breeding Bird Atlas, which is why this step is required
events_matrix <- mauricie_birds_df %>%
dplyr::select(SiteCode, survey_year, survey_month, survey_day, ObservationCount, latitude, longitude) %>%
distinct() %>%
sf::st_as_sf(coords = c("longitude", "latitude"), crs = 4326)Assign a point id identifier to each location based on its unique geometry and return the result as a dataframe.
events_matrix <- events_matrix %>%
group_by(SiteCode, geometry) %>%
mutate(point_id = cur_group_id()) %>%
ungroup()Restore the longitude and latitude columns, drop the geometry, convert to a dataframe, and keep only the relevant columns.
events_matrix <- events_matrix %>%
mutate(longitude = sf::st_coordinates(.)[,1], # Extract longitude
latitude = sf::st_coordinates(.)[,2]) %>% # Extract latitude
sf::st_drop_geometry() %>% # Drop geometry column
as.data.frame() %>%
dplyr::select(point_id, SiteCode, survey_year, survey_month, survey_day, longitude, latitude) # Keep relevant columnsJoin the environmental variables to the events matrix based on
point_id.
enviro_events_matrix <- left_join(events_matrix, enviro_data_df, by = "point_id")Filter the NatureCounts data for a species of interest and then merge the events matrix with the species-specific data. This will result in NA values for events where the species was not observed.
search_species("Golden-crowned Kinglet") #species_id = 14960
#> # A tibble: 1 × 5
#> species_id scientific_name english_name french_name taxon_group
#> <int> <chr> <chr> <chr> <chr>
#> 1 14960 Regulus satrapa Golden-crowned Kinglet Roitelet à couronne dorée BIRDS
kinglet_events <- mauricie_birds_df %>%
filter(species_id == "14960")
kinglet_events <- left_join(enviro_events_matrix, kinglet_events,
by = c("SiteCode",
"survey_year",
"survey_month",
"survey_day",
"latitude",
"longitude"))Zero-fill for specific species. Here, we replace the NA values in the
ObservationColumn with 0 and select the column of interest
to inspect your data.
kinglet_zero_fill <- kinglet_events %>%
mutate(ObservationCount = replace(ObservationCount, is.na(ObservationCount), 0)) %>% dplyr::select(point_id, SiteCode, survey_day, survey_month, survey_year, latitude, longitude, elevation:n_individuals, ObservationCount) Create a Presence/Absence column based on
ObservationCount.
kinglet_presence_absence <- kinglet_zero_fill %>%
mutate(Presence = ifelse(ObservationCount > 0, 1, 0)) # 1 = Presence, 0 = AbsenceCreate a bar plot to summarize the Presence/Absence data, grouped by
survey_year.
kinglet_PA_summary_year <- kinglet_presence_absence %>%
group_by(survey_year, category = ifelse(ObservationCount > 0, "Presence", "Absence")) %>%
summarise(count = n(), .groups = "drop")
ggplot(kinglet_PA_summary_year, aes(x = as.factor(survey_year), y = count, fill = category)) +
geom_col(position = "dodge") +
scale_fill_manual(values = c("Presence" = "darkgreen", "Absence" = "lightgray")) +
labs(title = "Golden-crowned Kinglet Presence/Absence per Survey Year",
x = "Survey Year",
y = "Number of Observations") +
theme_minimal()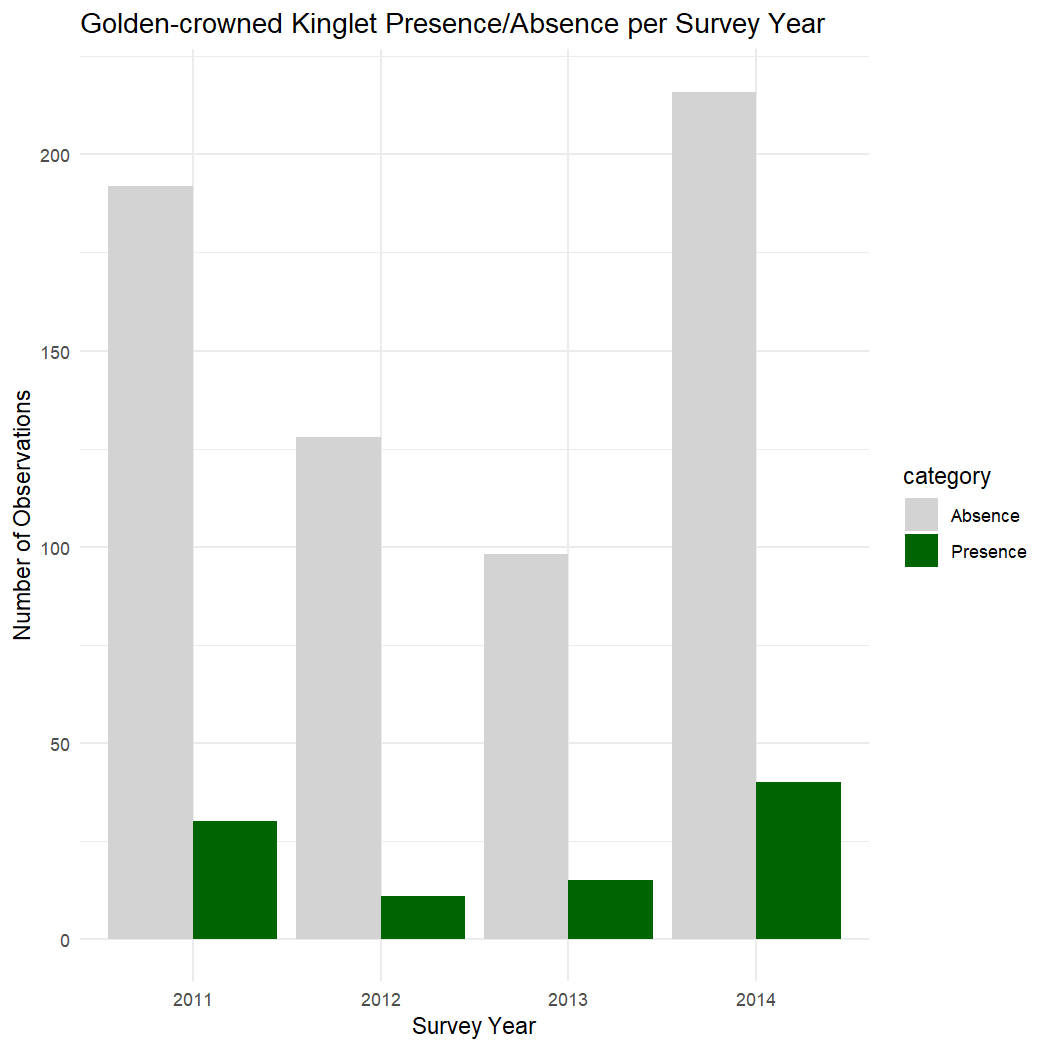
Now we can explore how Golden-crowned Kinglet presence/absence are related to environmental covariates.
First we will create a box plot for a single covariate.
ggplot(kinglet_presence_absence, aes(x = factor(Presence), y = ndvi, fill = factor(Presence))) +
geom_boxplot() +
labs(x = "Presence/Absence", y = "NDVI") +
ggtitle("Distribution of NDVI by Presence/Absence") +
theme_classic()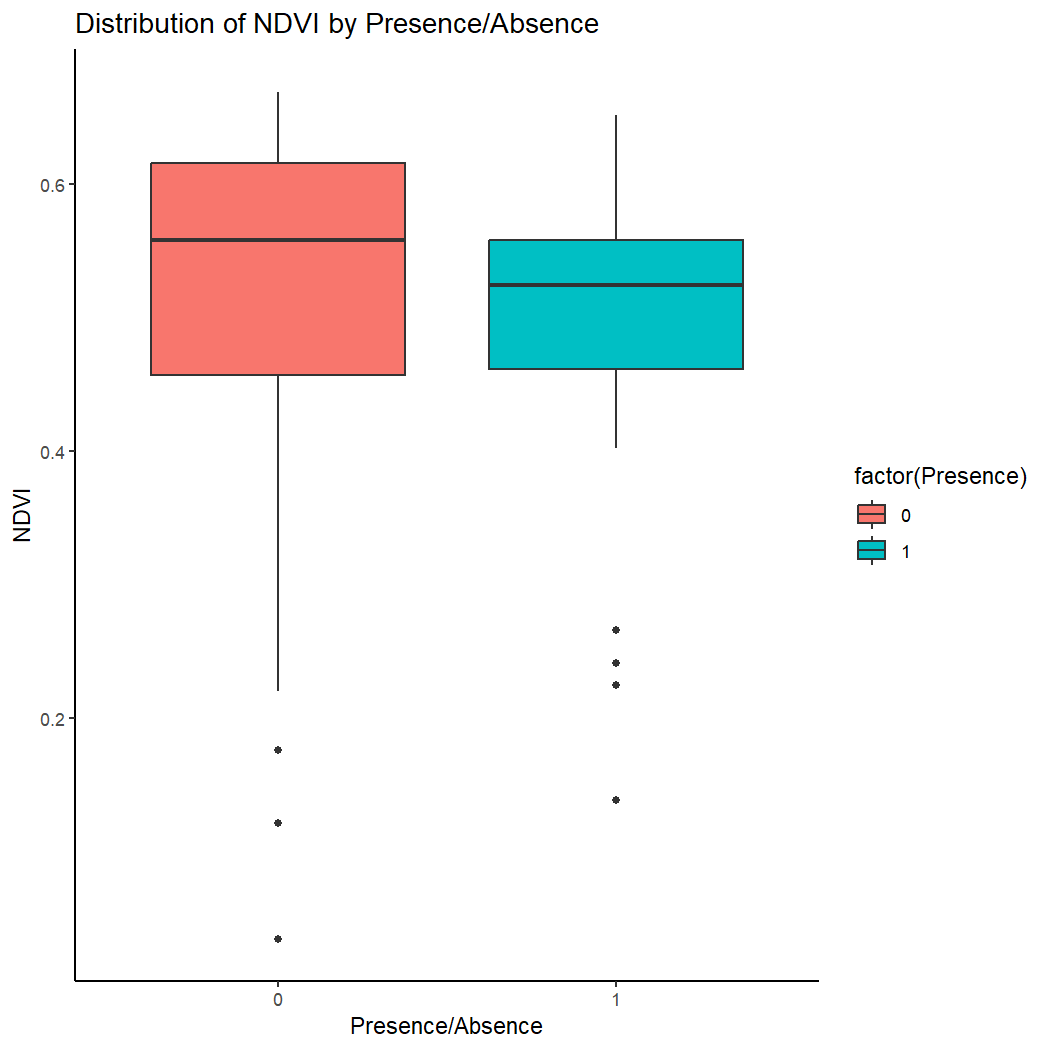
Or you can create a boxplot of multiple covariates using the pivot_longer function.
# Reshape data for plotting multiple covariates
kinglet_presence_absence_long <- pivot_longer(kinglet_presence_absence, cols = c("elevation", "ed", "pland_needleleaf", "pland_broadleaf", "pland_shrubland", "ndvi"), names_to = "Covariate", values_to = "Value")
# Create box plots for all covariates
ggplot(kinglet_presence_absence_long, aes(x = factor(Presence), y = Value, fill = factor(Presence))) +
geom_boxplot() +
facet_wrap(~ Covariate, scales = "free_y") +
labs(x = "Presence/Absence", y = "Covariate Value") +
ggtitle("Distribution of Covariates by Presence/Absence") +
theme(legend.position = "none")+
theme_classic()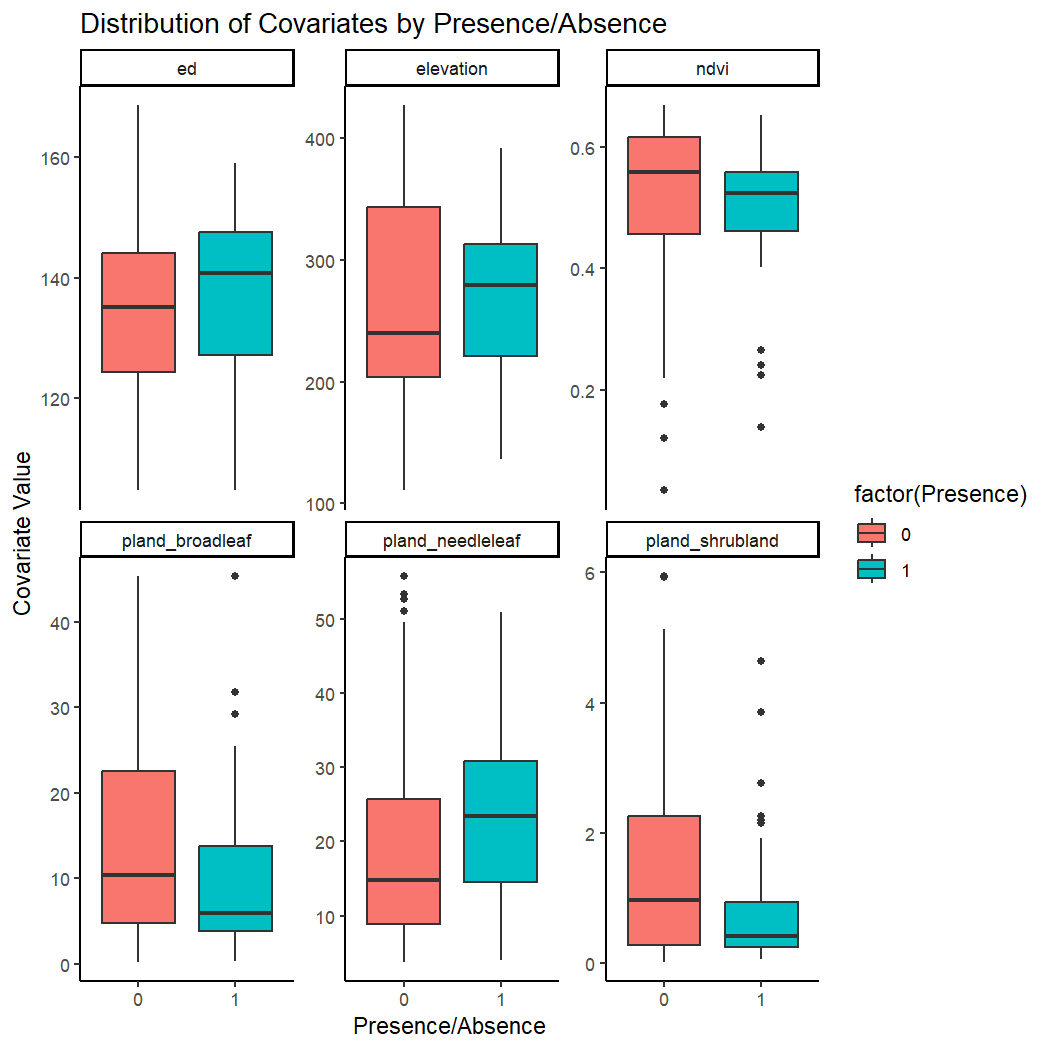
Users may wish to explore these data using simple statistics, like logistic regression, and create response curves showing the relationship between covariates and the probability of a species being present.
# Fit logistic regression model
model <- glm(Presence ~ ed + elevation + ndvi + pland_broadleaf + pland_needleleaf + pland_shrubland,
data = kinglet_presence_absence, family = binomial)
# Create sequence for covariate values
covar_range <- seq(min(kinglet_presence_absence$ed), max(kinglet_presence_absence$ed), length = 100)
# Generate predictions while holding other covariates at mean values
newdata <- data.frame(
ed = covar_range,
elevation = mean(kinglet_presence_absence$elevation),
ndvi = mean(kinglet_presence_absence$ndvi),
pland_broadleaf = mean(kinglet_presence_absence$pland_broadleaf),
pland_needleleaf = mean(kinglet_presence_absence$pland_needleleaf),
pland_shrubland = mean(kinglet_presence_absence$pland_shrubland)
)
pred_probs <- predict(model, newdata, type = "response")
# Plot response curve
plot(covar_range, pred_probs, type = "l",
xlab = "Edge Denisty", ylab = "Probability of Kinglet Presence",
main = "Response Curve for Edge Density")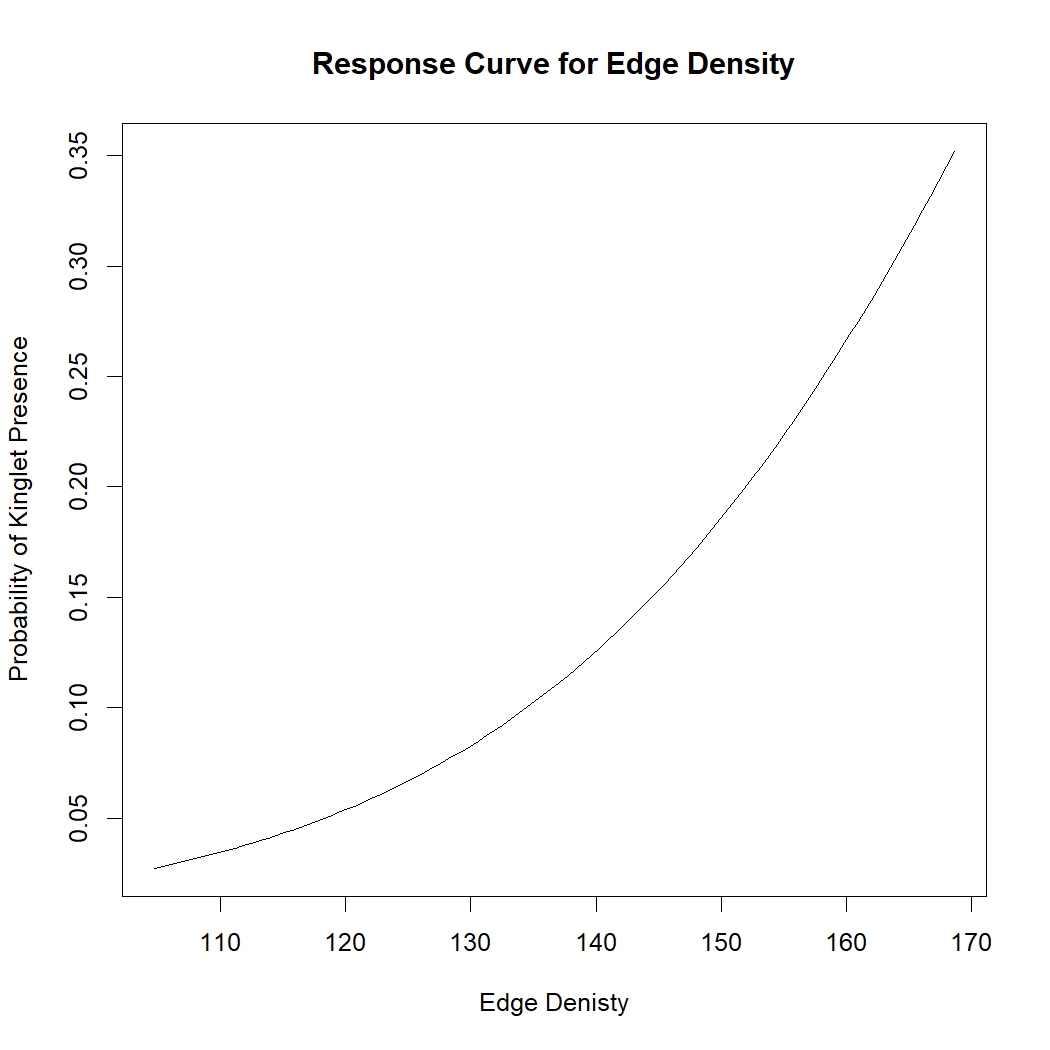 As edge density increase, the probability of a Golden Crown Kinglet
increase slightly. Users can explore these relationships for other
environmental covariates.
As edge density increase, the probability of a Golden Crown Kinglet
increase slightly. Users can explore these relationships for other
environmental covariates.
________________________________________________________________________________________________________________
Congratulations! You completed Chapter 7 - Summary Tools. In this chapter, you successfully summarized and visualized environmental data, biodiversity counts, and presence/absence data using various plotting techniques.