Chapter 1 - Spatial Data Exploration
2025-03-27
Source:vignettes/articles/2.1-SpatialDataExploration.Rmd
2.1-SpatialDataExploration.RmdChapter 1: Spatial Data Exploration
Authors: Dimitrios Markou, Danielle Ethier
naturecounts R package. It explains how to access,
view, filter, manipulate, and visualize NatureCounts data. We recommend
reviewing this tutorial before proceeding.1.0 Learning Objectives
By the end of Chapter 1 - Spatial Data Exploration, users will know how to:
Distinguish between types of spatial data (vector vs raster)
Select from a variety of geoprocessing functions in the
sfpackageVisualize NatureCounts data using spatio-temporal maps
This R tutorial requires the following packages:
1.1 Spatial Data Types
Spatial data is any type of vector or raster data that represents a feature or phenomena across geographic space.
The most common format used to store vector data in a file on disk is the ESRI Shapefile format (.shp). Shapefiles are always accompanied by files with .dbf, .shx, and .prj extensions.
Raster data files are typically stored with TIFF or GeoTIFF files with a (.tif) or (.tiff) extension. Raster data manipulation will be in covered in subsequent chapters (see Chapter 3: Climate Data, Chapter 4: Elevation Data, Chapter 5: Land Cover Data, Chapter 6: Satellite Imagery, and Chapter 7: Raster Summary).
Vector and raster data may also be associated with attribute data or temporal data. Attribute data provides additional information on the characteristics of spatial features while temporal data assigns a specific date or time range.
The sf package provides simple feature access in R.
This package works best with spatial data (point, line, polygon,
multipolygon) associated with tabular attributes (e.g shapefiles). You
may be familiar with the sp package that has similar
functionality in a different format, however, this package is no longer
in use as of 2023 and does not support integration with
tidyverse which is very popular among data scientists who
use R.
1.2 Geo-processing Functions
Geoprocessing functions allow us to manipulate or compute spatial
objects based on interactions between their geometries. There are
several useful functions integrated into the sf package
including:
st_transform() - transforms the CRS
of a specified CRS objectst_drop_geometry() - removes the geometry column of a sf
objectst_intersection(x, y) - creates geometry of the shared
portion of x and yst_crop(x, y, ..., xmin, ymin, xmax, ymax) - creates
geometry of x that intersects a specified rangest_difference(x, y) - creates geometry from x that does not
intersect with yst_area, st_length, and
st_distance can also be used to compute geometric
measurementsMore resources, including an sf package
cheatsheet can be found here.
1.3 Spatio-temporal Mapping
Example 1 - You would like to visualize the spatio-temporal distribution of Cedar Waxwing observations in June of each survey year using data from the Maritimes Breeding Bird Atlas (2006-2010).
Let’s fetch the NatureCounts data.
First, we look to find the collection code for the
Maritimes Breeding Bird Atlas.
collections <- meta_collections()
View(meta_collections())Second, we look to find the numeric species id.
search_species("cedar waxwing")
#> # A tibble: 2 × 5
#> species_id scientific_name english_name french_name taxon_group
#> <int> <chr> <chr> <chr> <chr>
#> 1 16330 Bombycilla cedrorum Cedar Waxwing Jaseur d'Amérique BIRDS
#> 2 40749 Bombycilla garrulus/cedrorum Bohemian/Cedar Waxwing Jaseur boréal ou J. d'Amérique BIRDSNow we can download the data.
The data download will not work unless you replace
"testuser"with your actual user name. You will be prompted to enter your password.
cedar_waxwing <- nc_data_dl(collections = "MBBA2PC", species = 16330,
username = "testuser", info = "spatial_data_tutorial")
#> Using filters: collections (MBBA2PC); species (16330); fields_set (BMDE2.00-min)
#> Collecting available records...
#> collection nrecords
#> 1 MBBA2PC 1659
#> Total records: 1,659
#>
#> Downloading records for each collection:
#> MBBA2PC
#> Records 1 to 1659 / 1659Use the format_dates function to create date and day-of-year (doy) columns.
cedar_waxwing <- format_dates(cedar_waxwing)Filter the data to only include observations from the month of June.
Convert the NatureCounts data to a spatial object using the point count coordinates.
cedar_waxwing_june_sf <- st_as_sf(cedar_waxwing_june,
coords = c("longitude", "latitude"), crs = 4326)Finally, use ggplot2 to visualize the spatio-temporal
distribution of Cedar Waxwing observations across the Maritime provinces
by color-coding the data points by survey_year and
creating a multi-panel plot based on this discrete variable:
ggplot(data = cedar_waxwing_june_sf) +
# Select a basemap
annotation_map_tile(type = "cartolight", zoom = NULL, progress = "none") +
# Plot the points, color-coded by survey_year
geom_sf(aes(color = as.factor(survey_year)), size = 1) +
# Facet by survey_year to create the multi-paneled map
facet_wrap(~ survey_year) +
# Customize the color scale
scale_color_brewer(palette = "Set1", name = "Survey Year") +
# Add a theme with a minimal design and change the font styles, to your preference
theme_minimal() +
theme(legend.position = "bottom") +
# To make the points in the legend larger without affecting map points
guides(color = guide_legend(override.aes = list(size = 3))) +
# Define the title and axis names
labs(title = "Cedar Waxwing June Observations by Survey Year",
x = "Longitude",
y = "Latitude")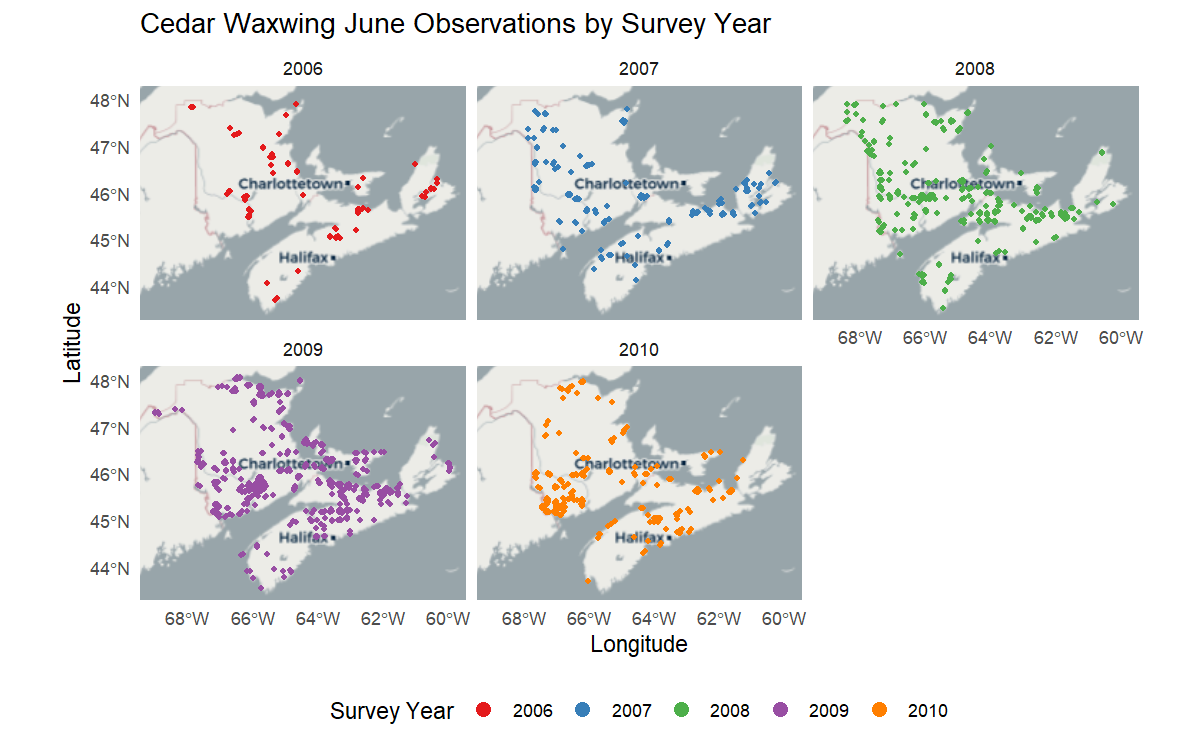
The map above provides a simple visualization of NatureCounts data over a broad spatial and temporal scale.
Congratulations! You completed Chapter 1: Spatial Data Exploration. Here, you successfully visualized NatureCounts vector data over a wide spatial and temporal scale using a multi-panel plot. In Chapter 2, you can explore spatial data manipulation, apply geoprocessing functions, and visualize NatureCounts data within Key Biodiversity Areas (KBAs) and Priority Places.