Chapter 3 - Climate Data
2025-04-02
Source:vignettes/articles/2.3-ClimateData.Rmd
2.3-ClimateData.RmdChapter 3: Climate Data
Authors: Dimitrios Markou, Danielle Ethier
3.0 Learning Objectives
By the end of Chapter 3 - Climate Data, users will know how to:
- Download and preprocess vector and raster climate data
- Combine NatureCounts observations with climate data
- Visualize NatureCounts and climate data using plots and spatio-temporal maps
This tutorial utilizes the following bird occurrence, spatial, and climate data sources:
| Data | Description |
|---|---|
| eBird Canada (Prairies) | NatureCounts, EBIRD-CA-PR (1800-2024) |
| Alberta Breeding Bird Atlas | NatureCounts, ABATLAS1 (1987-1992) and ABATLAS2 (2000-2005) |
| Alberta Bird Records | NatureCounts, ABIRDRECS (1941-2006) |
| Whooping Crane Nesting Area and Summer Range | Key Biodiversity Area boundary (.shp) and site attributes |
| Environment and Climate Change Canada | Historical (vector) weather data, accessed through the
weathercan R package |
| WorldClim | Historical (raster) climate data. Includes nineteen bioclimatic variables representing the 1970-2000 average |
This tutorial requires the following packages:
library(naturecounts)
library(sf)
library(lubridate)
library(dplyr)
library(ggplot2)
library(terra)
library(leaflet)
# weathercan is in the R-Universe rather than on CRAN,
# so we install the package a little differently
#
# install.packages("weathercan",
# repos = c("https://ropensci.r-universe.dev",
# "https://cloud.r-project.org"))
library(weathercan)3.1 Data Setup
collections <- meta_collections()
View(meta_collections())
search_species("whooping crane")
View(search_species("whooping crane"))To download the NatureCounts data, you can specify the collection and
species code relevant to your research. Replace testuser
with your user name.
The data download will not work unless you replace
"testuser"with your actual user name. You will be prompted to enter your password.
whooping_crane_data <- nc_data_dl(
collections = c("ABATLAS1", "ABATLAS2", "ABBIRDRECS"), species = 4030,
username = "testuser", info = "spatial_data_tutorial")eBird has the greatest number of Whooping Crane records, however, this collection comprise data of Access Level 4. If you wish to access this collection you must sign up for a free account and make a data request. Otherwise, you can carry forward with the tutorial without these data and skip this code chunk.
whooping_crane_data <- nc_data_dl(
collections = c("ABATLAS1", "ABATLAS2", "ABBIRDRECS","EBIRD-CA-PR"), species = 4030,
username = "testuser", info = "spatial_data_tutorial")Grouping the NatureCounts data might provide more meaningful insight
on species occurrences at each site and how they might vary across time
and space. You can achieve this by summarizing species occurrence by
SiteCode and survey year, while keeping the coordinate information for
each site using the group_by and summarise
functions:
species_occurrence_summary <- whooping_crane_data %>%
format_dates() %>%
group_by(SiteCode, survey_year, longitude, latitude) %>%
summarise(total_count = sum(as.numeric(ObservationCount), na.rm = TRUE)) %>%
ungroup()
#> `summarise()` has grouped output by 'SiteCode', 'survey_year', 'longitude'. You can override using the `.groups` argument.Lastly, you can summarize the NatureCounts data once more to get the
annual total counts of Whooping Cranes, say between 1985 and 2011. To do
so, we can 1) produce a regular dataframe called
cranes_summary by dropping the geometry column
(st_drop_geometry) 2) calculate the
annual_count by using the summarise and
sum functions and 3) filter for NA values and by
year:
cranes_summary <- species_occurrence_summary %>%
st_drop_geometry() %>%
group_by(survey_year) %>%
summarize(annual_count = sum(total_count, na.rm = TRUE)) %>%
filter(!is.na(survey_year)) %>%
filter(survey_year >= 1985 & survey_year <= 2011)
cranes_summary
#> # A tibble: 12 × 2
#> survey_year annual_count
#> <int> <dbl>
#> 1 1987 0
#> 2 1988 0
#> 3 1989 0
#> 4 1990 0
#> 5 1991 0
#> 6 1996 1
#> 7 2000 24
#> 8 2001 14
#> 9 2002 26
#> 10 2003 14
#> 11 2004 18
#> 12 2005 1803.2 weathercan (Vector) Data
weathercan is an R package designed to help access
historical weather data from ECCC. Steffi LaZerte’s online
documentation, provides more details on how to download, filter, and
visualize weather data in R.
First, let’s take a look at the built in stations dataset:
stations()
#> The stations data frame hasn't been updated in over 4 weeks. Consider running `stations_dl()` to check for updates and make sure you have the most recent stations list available
#> # A tibble: 26,382 × 17
#> prov station_name station_id climate_id WMO_id TC_id lat lon elev tz interval start end normals normals_1991_2020
#> <chr> <chr> <dbl> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 AB DAYSLAND 1795 301AR54 NA <NA> 52.9 -112. 689. Etc/G… day 1908 1922 FALSE FALSE
#> 2 AB DAYSLAND 1795 301AR54 NA <NA> 52.9 -112. 689. Etc/G… hour NA NA FALSE FALSE
#> 3 AB DAYSLAND 1795 301AR54 NA <NA> 52.9 -112. 689. Etc/G… month 1908 1922 FALSE FALSE
#> 4 AB EDMONTON CORONATION 1796 301BK03 NA <NA> 53.6 -114. 671. Etc/G… day 1978 1979 FALSE FALSE
#> 5 AB EDMONTON CORONATION 1796 301BK03 NA <NA> 53.6 -114. 671. Etc/G… hour NA NA FALSE FALSE
#> 6 AB EDMONTON CORONATION 1796 301BK03 NA <NA> 53.6 -114. 671. Etc/G… month 1978 1979 FALSE FALSE
#> 7 AB FLEET 1797 301B6L0 NA <NA> 52.2 -112. 838. Etc/G… day 1987 1990 FALSE FALSE
#> 8 AB FLEET 1797 301B6L0 NA <NA> 52.2 -112. 838. Etc/G… hour NA NA FALSE FALSE
#> 9 AB FLEET 1797 301B6L0 NA <NA> 52.2 -112. 838. Etc/G… month 1987 1990 FALSE FALSE
#> 10 AB GOLDEN VALLEY 1798 301B8LR NA <NA> 53.2 -110. 640 Etc/G… day 1987 1998 FALSE FALSE
#> # ℹ 26,372 more rows
#> # ℹ 2 more variables: normals_1981_2010 <lgl>, normals_1971_2000 <lgl>You are ready to download weather data! For the purpose of this tutorial, let’s perform a smaller weather data download by specifying specific station IDs. The weather stations closest to Wood Buffalo National Park are BIRCH MOUNTAIN LO (station_id = 2481) and BUCKTON LO (station_id = 2486). These stations were identified using the the ECCC search option which helps you search for historical weather data based on proximity to a city, coordinate, or National Park.
birch_station <- stations_search(name = "BIRCH MOUNTAIN LO", interval = "day")
buckton_station <- stations_search(name = "BUCKTON LO", interval = "day")
wood_buff_stations <- rbind(birch_station, buckton_station) # combine Now you can perform the weather data download (give it a few minutes).
weather <- weather_dl(station_ids = wood_buff_stations$station_id,
start = "1985-01-01",
end = "2011-12-31",
interval = "day", quiet = TRUE)NOTE - Larger data downloads can be performed based on filtered stations datasets as well as start, end, and interval specifications. Large historical weather downloads can take a while!
Here is an example where we filtered based on province, interval, station operation period, and elevation to exclude extreme measurements in the weather data download:
To execute this code chunk, remove the #
#prov_stations <- stations %>%
# filter(prov %in% c("AB"),
# interval == "day",
# start >= 1960,
# end <= 2023,
# elev < 1000)To execute this code chunk, remove the #
# weather <- weather_dl(station_ids = prov_stations$station_id,
# start = "2000-01-01",
# end = "2023-12-31",
# interval = "day", quiet = TRUE)Finally, to summarize the weather data by calculating the annual mean
summer temperature and precipitation we can use the familiar
filter, group_by, and summarise
functions:
yearly_climate <- weather %>%
filter(month(date) %in% 5:8) %>% # Filter for summer months May (5) through August (8)
group_by(survey_year = year(date)) %>% # Group by year
summarize(
yearly_avg_temp = mean(mean_temp, na.rm = TRUE),
yearly_avg_precip = mean(total_precip, na.rm = TRUE)
)
yearly_climate
#> # A tibble: 27 × 3
#> survey_year yearly_avg_temp yearly_avg_precip
#> <dbl> <dbl> <dbl>
#> 1 1985 11.6 1.14
#> 2 1986 11.7 2.96
#> 3 1987 11.8 2.71
#> 4 1988 12.1 3.40
#> 5 1989 13.0 1.85
#> 6 1990 13.1 2.44
#> 7 1991 13.4 3.40
#> 8 1992 11.2 2.40
#> 9 1993 10.7 3.00
#> 10 1994 13.2 3.39
#> # ℹ 17 more rowsNow that you have prepared the NatureCounts and weathercan climate data you can combine the data together for analysis.
cranes_climate_summary <- left_join(cranes_summary, yearly_climate, by = "survey_year")
cranes_climate_summary
#> # A tibble: 12 × 4
#> survey_year annual_count yearly_avg_temp yearly_avg_precip
#> <dbl> <dbl> <dbl> <dbl>
#> 1 1987 0 11.8 2.71
#> 2 1988 0 12.1 3.40
#> 3 1989 0 13.0 1.85
#> 4 1990 0 13.1 2.44
#> 5 1991 0 13.4 3.40
#> 6 1996 1 11.7 3.37
#> 7 2000 24 10.6 3.83
#> 8 2001 14 13.7 2.76
#> 9 2002 26 11.3 3.18
#> 10 2003 14 12.9 2.72
#> 11 2004 18 10.6 2.08
#> 12 2005 180 10.7 2.89To visualize average annual summer temperature and total count, you can use a scatterplot.
ggplot(cranes_climate_summary, aes(x = yearly_avg_temp, y = annual_count)) +
geom_point(color = "blue", size = 3, alpha = 0.7) + # Simple blue points
geom_smooth(method = "lm", se = FALSE, color = "red") + # Add linear regression line
theme_minimal() +
labs(x = "Avg Temperature (°C)",
y = "Total Count",
title = "Whooping Crane Count vs. Annual Avg Summer Temp") +
theme(plot.title = element_text(hjust = 0.5)) # Center the title
#> `geom_smooth()` using formula = 'y ~ x'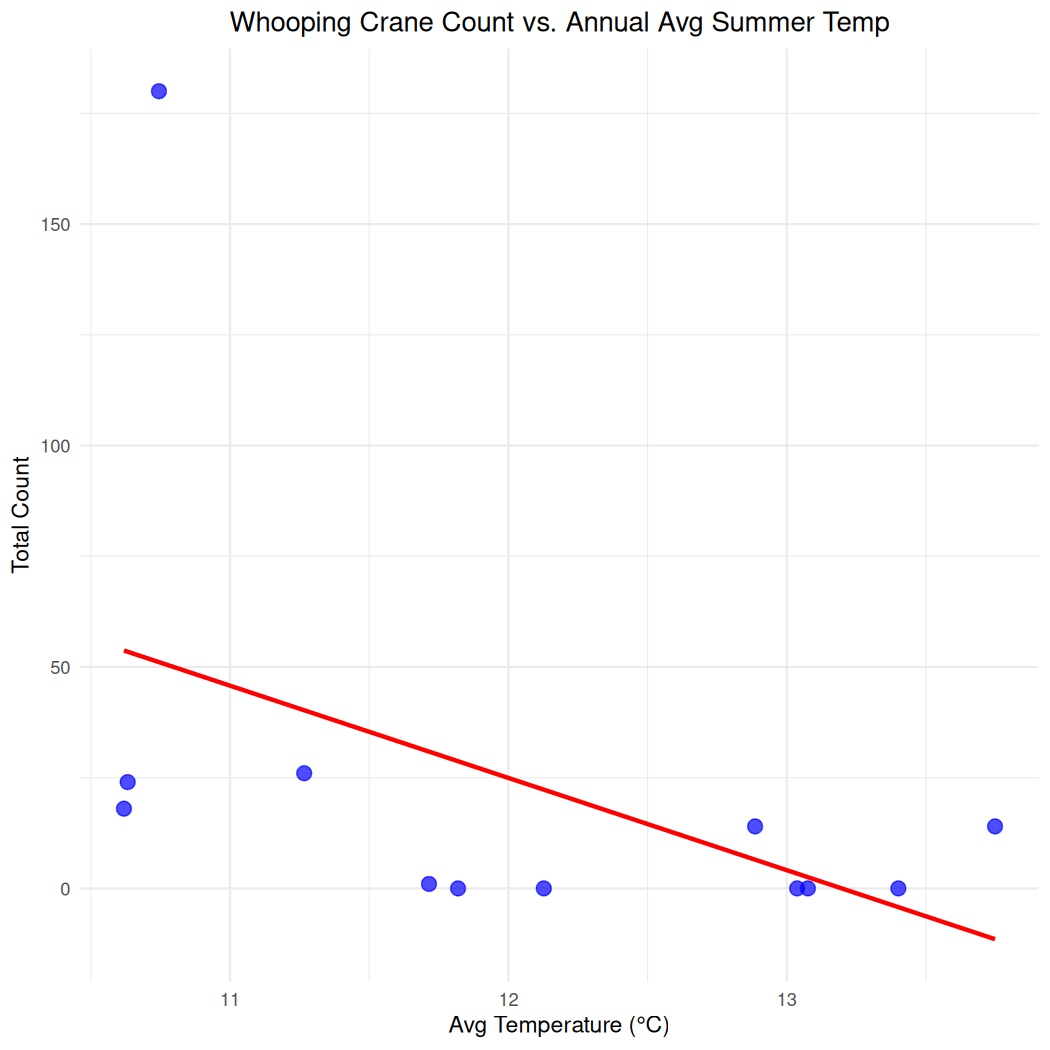
And do the same for annual summer precipitation:
ggplot(cranes_climate_summary, aes(x = yearly_avg_precip, y = annual_count)) +
geom_point(color = "blue", size = 3, alpha = 0.7) + # Simple blue points
geom_smooth(method = "lm", se = FALSE, color = "red") + # Add linear regression line
theme_minimal() +
labs(x = "Avg Precipitation",
y = "Total Count",
title = "Whooping Crane Count vs. Annual Avg Summer Precip") +
theme(plot.title = element_text(hjust = 0.5)) # Center the title
#> `geom_smooth()` using formula = 'y ~ x'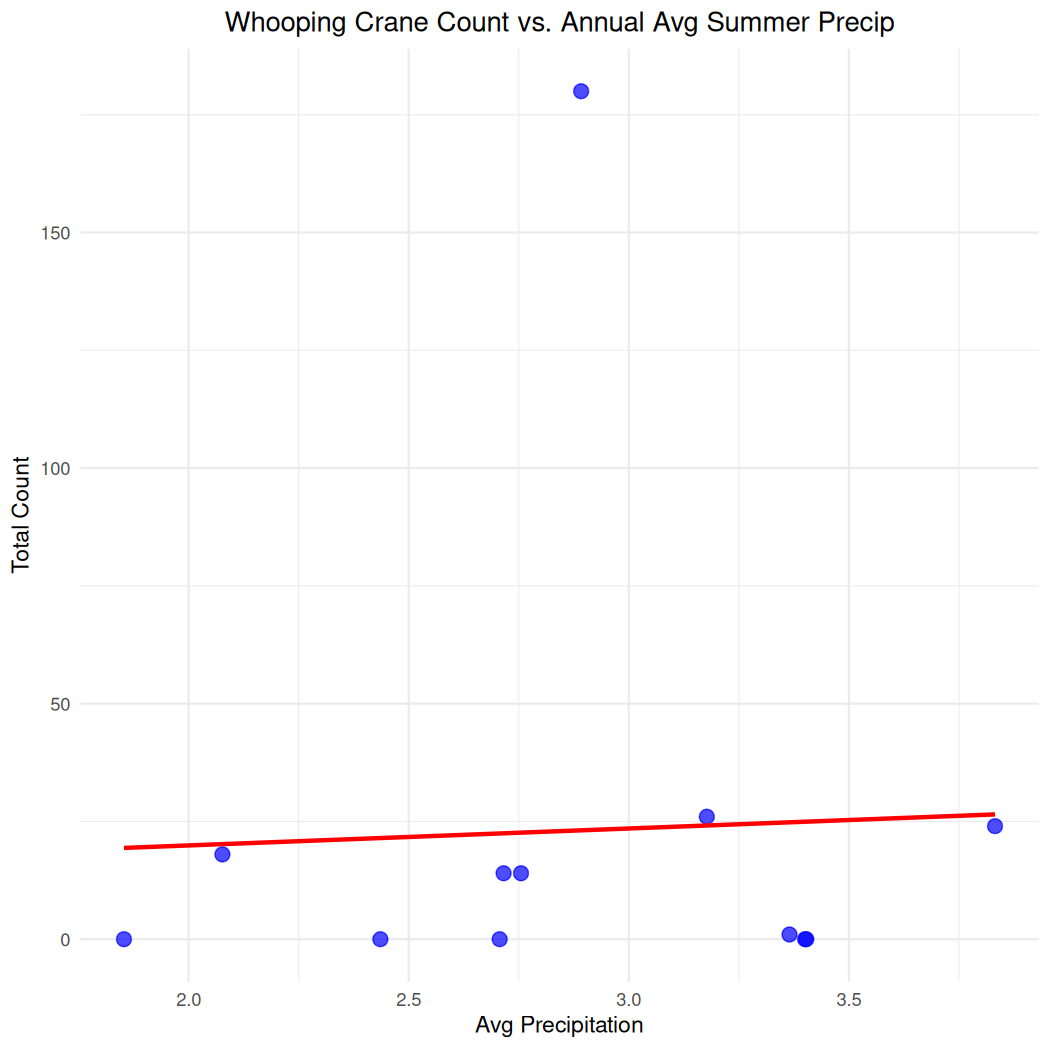
3.3 WorldClim (Raster) Data
WorldClim provides high spatial resolution global weather and climate data. Their data is in raster GeoTiff (.tif) file format and comprises a variety of variables including 19 bioclimatic variables representing annual trends, seasonality, and extreme environmental factors.
Download the bioclimatic data on this webpage. The data are available at the four spatial resolutions, between 30 seconds (~1 km2) to 10 minutes (~340 km2). Each download is a “zip” file containing 19 GeoTiff (.tif) files, one for each month of the variables. For this tutorial, we recommend you download the 10 minute resolution.
After downloading the WorldClim data to your working directory, list
all the files in the raster stack by specifying the path to your folder
(use list.files function) and read them into R using the
rast function.
This code will get the path to your working directory.
getwd()Now point the list.file to this directory using the
sample code below.
worldclim_list <- list.files(path = "YOUR/PATH/HERE", pattern = "\\.tif$", full.names = TRUE)
# Read and stack the raster files
worldclim_stack <- rast(worldclim_list)
# Print information about the stack
worldclim_stack#> class : SpatRaster
#> dimensions : 1080, 2160, 19 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> sources : wc2.1_10m_bio_1.tif
#> wc2.1_10m_bio_10.tif
#> wc2.1_10m_bio_11.tif
#> ... and 16 more sources
#> names : wc2.1~bio_1, wc2.1~io_10, wc2.1~io_11, wc2.1~io_12, wc2.1~io_13, wc2.1~io_14, ...
#> min values : -54.72435, -37.78142, -66.31125, 0, 0, 0, ...
#> max values : 30.98764, 38.21617, 29.15299, 11191, 2381, 484, ...Great! The raster stack which includes the 19 bio-climatic variables has been read into R successfully. We can simplify the layer names which represent each variable, respectively:
Let’s take a look at the first layer ‘bio1’ which is Annual Mean Temperature. To do so, you can subset the layers of the SpatRaster with $ or two sets of square brackets [ [ ] ].
# Plot the first bio-climatic layer (e.g., bio1: Annual Mean Temperature)
plot(worldclim_stack[["bio1"]], main = "Annual Mean Temperature (bio1)")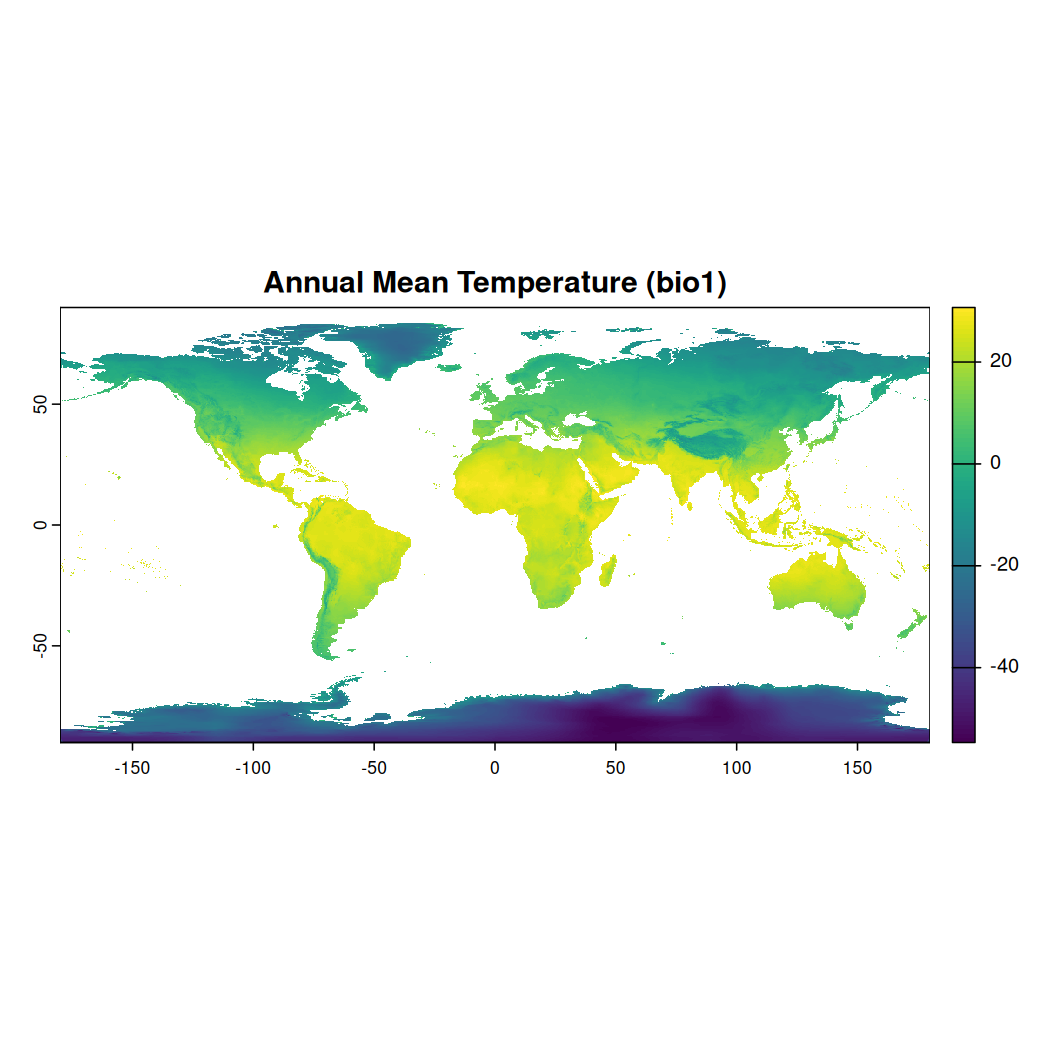
# Summary statistics for each layer
summary(worldclim_stack)
#> Warning: [summary] used a sample
#> bio1 bio2 bio3 bio4 bio5 bio6 bio7
#> Min. :-54.72 Min. :-37.72 Min. :-66.19 Min. : 0.0 Min. : 0.0 Min. : 0.00 Min. : 0.00
#> 1st Qu.:-22.81 1st Qu.:-10.58 1st Qu.:-34.22 1st Qu.: 110.0 1st Qu.: 28.0 1st Qu.: 0.00 1st Qu.: 41.41
#> Median : -0.53 Median : 13.82 Median :-15.20 Median : 337.0 Median : 60.0 Median : 5.00 Median : 64.80
#> Mean : -4.05 Mean : 7.19 Mean :-13.90 Mean : 550.3 Mean : 93.5 Mean : 15.39 Mean : 74.63
#> 3rd Qu.: 19.02 3rd Qu.: 24.80 3rd Qu.: 12.13 3rd Qu.: 690.0 3rd Qu.: 112.0 3rd Qu.: 18.00 3rd Qu.:100.13
#> Max. : 30.71 Max. : 38.18 Max. : 28.62 Max. :7011.0 Max. :1965.0 Max. :471.00 Max. :228.88
#> NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591
#> bio8 bio9 bio10 bio11 bio12 bio13 bio14
#> Min. : 0.0 Min. : 0.00 Min. : 0.0 Min. : 0.0 Min. : 1.00 Min. : 9.65 Min. : 0.0
#> 1st Qu.: 56.0 1st Qu.: 2.00 1st Qu.: 7.0 1st Qu.: 17.0 1st Qu.: 7.38 1st Qu.: 20.97 1st Qu.: 569.8
#> Median : 152.0 Median : 23.00 Median : 107.0 Median : 47.0 Median : 9.34 Median : 26.57 Median : 897.1
#> Mean : 241.8 Mean : 55.47 Mean : 156.5 Mean : 108.8 Mean : 9.94 Mean : 34.54 Mean : 880.0
#> 3rd Qu.: 298.0 3rd Qu.: 66.00 3rd Qu.: 225.0 3rd Qu.: 109.0 3rd Qu.:12.36 3rd Qu.: 45.35 3rd Qu.:1206.6
#> Max. :4503.0 Max. :1477.00 Max. :3159.0 Max. :3849.0 Max. :20.28 Max. :100.00 Max. :2342.4
#> NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591
#> bio15 bio16 bio17 bio18 bio19
#> Min. :-29.60 Min. :-72.40 Min. : 1.00 Min. :-66.19 Min. :-54.26
#> 1st Qu.: -6.01 1st Qu.:-39.30 1st Qu.:26.11 1st Qu.:-26.12 1st Qu.:-24.25
#> Median : 21.53 Median :-21.61 Median :33.38 Median : 10.69 Median : -9.26
#> Mean : 13.93 Mean :-19.80 Mean :33.72 Mean : -0.93 Mean : -5.38
#> 3rd Qu.: 31.35 3rd Qu.: 4.63 3rd Qu.:41.31 3rd Qu.: 21.46 3rd Qu.: 18.52
#> Max. : 47.95 Max. : 26.20 Max. :71.58 Max. : 37.70 Max. : 37.04
#> NA's :65591 NA's :65591 NA's :65591 NA's :65591 NA's :65591Convert the species_occurrences_summary, which
summarizes species occurrence by SiteCode, into a spatial object using
the st_as_sf function:
Extract the bio-climatic variable values for each bird observation site, respectively.
bioclim_values <- terra::extract(worldclim_stack, cranes_sf)Then combine the bio-climatic values with the NatureCounts data.
bioclim_data <- cbind(cranes_sf, bioclim_values,
longitude = species_occurrence_summary$longitude,
latitude = species_occurrence_summary$latitude)Filter the data to only include observations made post 1970 to match the temporal resolution of the climate dataset:
bioclim_data <- bioclim_data %>%
filter(survey_year >= "1970")
summary(bioclim_data)
#> SiteCode survey_year total_count ID bio1 bio2 bio3
#> Length:67 Min. :1987 Min. : 0.000 Min. : 1.0 Min. :-2.7267 Min. :14.11 Min. :-21.910
#> Class :character 1st Qu.:2000 1st Qu.: 1.000 1st Qu.:17.5 1st Qu.:-2.4017 1st Qu.:14.73 1st Qu.:-21.183
#> Mode :character Median :2002 Median : 1.000 Median :34.0 Median :-2.3429 Median :14.80 Median :-20.766
#> Mean :2000 Mean : 4.134 Mean :34.0 Mean :-0.6792 Mean :14.99 Mean :-17.939
#> 3rd Qu.:2003 3rd Qu.: 2.000 3rd Qu.:50.5 3rd Qu.: 1.3970 3rd Qu.:15.31 3rd Qu.:-14.532
#> Max. :2005 Max. :160.000 Max. :67.0 Max. : 4.0880 Max. :15.83 Max. : -7.527
#> bio4 bio5 bio6 bio7 bio8 bio9 bio10 bio11
#> Min. :346.0 Min. : 52.0 Min. :10.00 Min. :42.27 Min. :135.0 Min. :34.00 Min. :134.0 Min. :34.00
#> 1st Qu.:352.0 1st Qu.: 56.0 1st Qu.:14.00 1st Qu.:46.05 1st Qu.:145.0 1st Qu.:44.00 1st Qu.:145.0 1st Qu.:51.50
#> Median :354.0 Median : 56.0 Median :14.00 Median :46.59 Median :147.0 Median :44.00 Median :147.0 Median :53.00
#> Mean :390.0 Mean : 67.9 Mean :13.88 Mean :54.81 Mean :178.3 Mean :46.51 Mean :178.3 Mean :52.15
#> 3rd Qu.:425.5 3rd Qu.: 80.5 3rd Qu.:14.00 3rd Qu.:65.93 3rd Qu.:215.0 3rd Qu.:49.50 3rd Qu.:215.0 3rd Qu.:53.00
#> Max. :543.0 Max. :104.0 Max. :17.00 Max. :74.12 Max. :270.0 Max. :56.00 Max. :270.0 Max. :61.00
#> bio12 bio13 bio14 bio15 bio16 bio17 bio18 bio19
#> Min. :10.98 Min. :21.53 Min. : 940.5 Min. :21.38 Min. :-29.17 Min. :39.16 Min. :12.90 Min. :-13.17
#> 1st Qu.:11.82 1st Qu.:23.42 1st Qu.:1214.9 1st Qu.:22.73 1st Qu.:-28.41 1st Qu.:44.32 1st Qu.:14.73 1st Qu.:-11.26
#> Median :12.06 Median :23.86 Median :1443.2 Median :22.96 Median :-27.86 Median :50.79 Median :14.80 Median :-10.89
#> Mean :11.99 Mean :25.17 Mean :1342.1 Mean :22.83 Mean :-25.21 Mean :48.04 Mean :14.96 Mean :-10.78
#> 3rd Qu.:12.07 3rd Qu.:26.12 3rd Qu.:1465.4 3rd Qu.:23.06 3rd Qu.:-21.73 3rd Qu.:51.47 3rd Qu.:15.31 3rd Qu.:-10.72
#> Max. :13.89 Max. :33.67 Max. :1499.5 Max. :24.50 Max. :-15.46 Max. :51.83 Max. :15.83 Max. : -3.01
#> longitude latitude geometry
#> Min. :-117.1 Min. :51.05 POINT :67
#> 1st Qu.:-113.3 1st Qu.:54.24 epsg:4326 : 0
#> Median :-113.0 Median :59.90 +proj=long...: 0
#> Mean :-112.7 Mean :57.49
#> 3rd Qu.:-111.7 3rd Qu.:59.97
#> Max. :-110.2 Max. :60.21Visualize the distribution of bio1 compared to total_count:
# Scatter plot of bird observations against a bioclimatic variable (bio1 = Annual Mean Temperature)
ggplot(data = bioclim_data, aes(x = bio1, y = total_count)) +
geom_point(alpha = 0.6) +
labs(title = "Bird Observations vs. Bioclimatic Variable (bio1)",
x = "Annual Mean Temperature",
y = "ObservationCount") +
theme_minimal()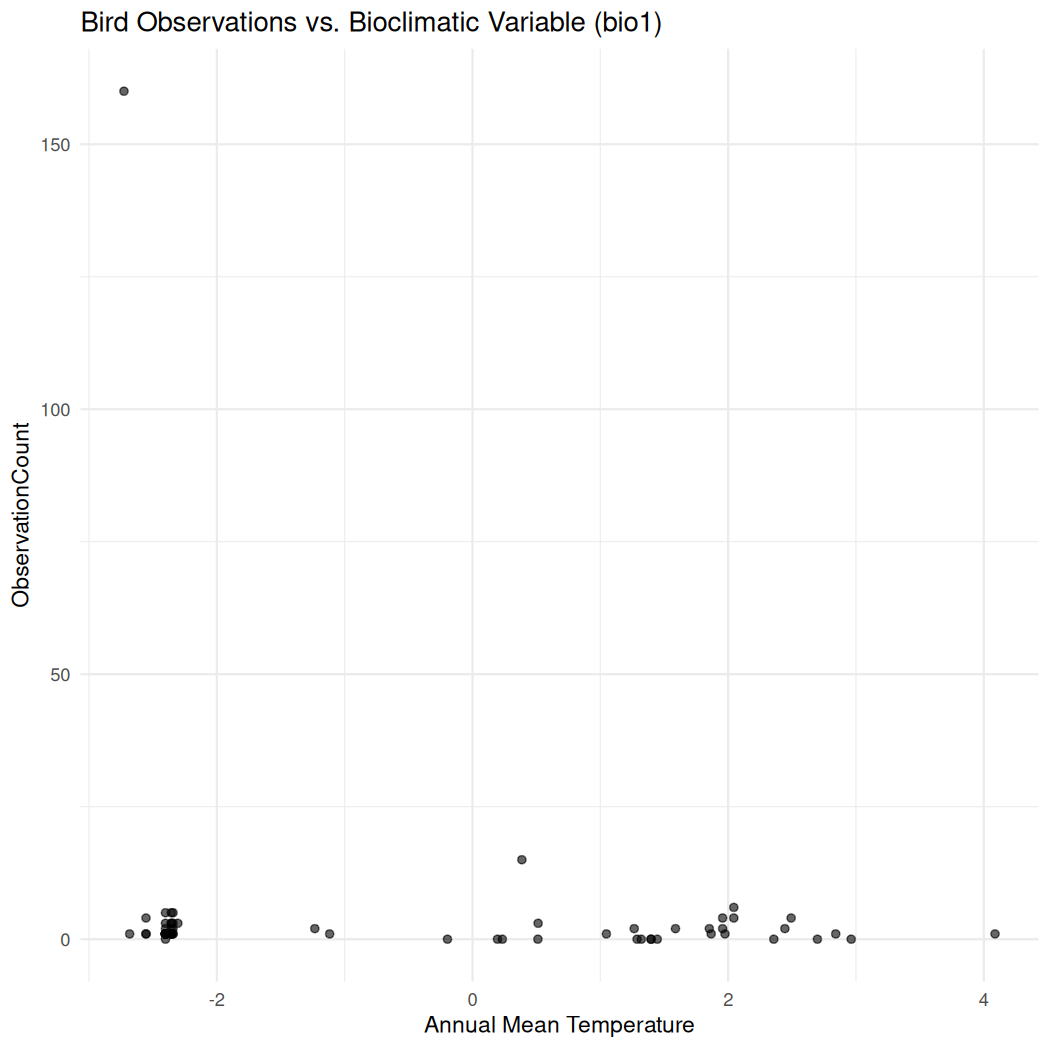
Map the NatureCounts data by grouping it by SiteCode, sizing the points by total_count and colorizing them by a bio-climatic variable (bio1):
bioclim_data <- st_transform(bioclim_data, crs = 4326)To make an interactive map of total counts per site.
# Group by SiteCode and summarize total_count
bioclim_data <- bioclim_data %>%
group_by(SiteCode) %>%
summarize(total_count = sum(total_count, na.rm = TRUE),
bio1 = mean(bio1, na.rm = TRUE),
latitude = first(latitude),
longitude = first(longitude))
# Define color palette for bio1
pal <- colorNumeric(palette = "YlOrRd", domain = bioclim_data$bio1)
# Define a scaling factor for total_count
scaling_factor <- 2 # Adjust this to scale the circle size
# Plot using leaflet
leaflet(bioclim_data) %>%
addTiles() %>%
addCircleMarkers(
~longitude, ~latitude,
radius = ~pmin(pmax(sqrt(total_count) * scaling_factor, 3), 15), # Scale and limit radius
fillColor = ~pal(bio1), # Color points by bio1
color = "black", stroke = TRUE, fillOpacity = 0.8,
popup = ~paste0("<strong>SiteCode:</strong> ", SiteCode, "<br>",
"<strong>Total Count:</strong> ", total_count, "<br>",
"<strong>Bio1:</strong> ", bio1) # creates info popup labels
) %>%
addLegend(pal = pal, values = ~bio1, title = "Bio1", position = "bottomright") %>%
addScaleBar(position = "bottomleft", options = scaleBarOptions(imperial = FALSE))Congratulations! You’ve completed Chapter 3 - Climate Data. Here, you successfully combined NatureCounts data with vector and raster climate data in R. In Chapter 4 you can explore how-to link NatureCounts observations with elevation data.